
Friedrich-Alexander-Universität Erlangen
Lehrstuhl für Mustererkennung
Martensstraße 3
91058 Erlangen

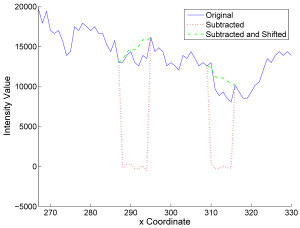 |
High-density objects, like catheters, pacemakers or even contrast agent-filled vessels, cause characteristic streak artifacts in computed tomography (CT). Similar to metal artifacts, these streaks can be reduced by removing the dense object using segmentation and interpolation. First, we compare state-of-the-art interpolation methods like linear, spline and higher-order methods to the Healing Brush technique. Second, a new method is presented, that extracts a low-frequency model of the dense object and restores the decomposed X-ray intensity of the remaining tissue. This method is henceforth called Subtract-and-Shift. Compared to standard interpolation methods, it retains the measured structure that is superimposed and dominated by the dense object. The extracted structure is then used to replace the segmented pixel intensities of the object. The introduced method is compared to state-of-the-art interpolation methods using in-vivo data. First preliminary results show that Subtract-and-Shift can be superior to these interpolation methods.
High-density objects, like catheters, pacemakers or even contrast agent-filled vessels, cause characteristic streak artifacts in computed tomography (CT). Similar to metal artifacts, these streaks can be reduced by removing the dense object using segmentation and interpolation. First, we compare state-of-the-art interpolation methods like linear, spline and higher-order methods to the Healing Brush technique. Second, a new method is presented, that extracts a low-frequency model of the dense object and restores the decomposed X-ray intensity of the remaining tissue. This method is henceforth called Subtract-and-Shift. Compared to standard interpolation methods, it retains the measured structure that is superimposed and dominated by the dense object. The extracted structure is then used to replace the segmented pixel intensities of the object. The introduced method is compared to state-of-the-art interpolation methods using in-vivo data. First preliminary results show that Subtract-and-Shift can be superior to these interpolation methods.
 |
 |
 |
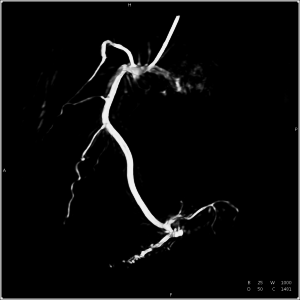
Generating 3-D reconstructions of cardiac vasculature from angiographic C-arm CT (rotational angiography) data is a challenging problem. Currently, many approaches depend on a reconstruction from ECG-gated projection data either as a reference for further processing or as the final result. Due to imperfect gating, e.g. caused by irregular heart movement, residual motion corrupts these reconstructions. We present an algorithm to compensate for this residual motion. The approach is based on a deformable 2-D–2-D registration between the acquired projection data and a forward projection of the initial ECG-gated reconstruction. It does not depend on an explicit segmentation of vasculature or markers, and works without user interaction. The estimated 2-D deformation field is compensated for in the backprojection step of a subsequent reconstruction. The algorithm is evaluated on six clinical datasets, showing a clear decrease in artefact level and better visibility of structure in the compensated reconstructions.
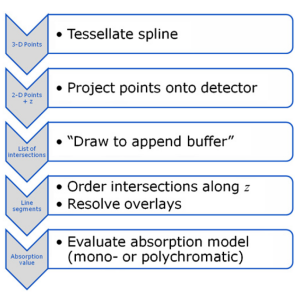
Many scientists in the field of x-ray imaging rely on the simulation of x-ray images. As the phantom models become more and more realistic, their projection requires high computational effort. Since x-ray images are based on transmission, many standard graphics acceleration algorithms cannot be applied to this task. However, if adapted properly, the simulation speed can be increased dramatically using state-of-the-art graphics hardware. A custom graphics pipeline that simulates transmission projections for tomographic reconstruction was implemented based on moving spline surface models. All steps from tessellation of the splines, projection onto the detector and drawing are implemented in OpenCL. We introduced a special append buffer for increased performance in order to store the intersections with the scene for every ray. Intersections are then sorted and resolved to materials. Lastly, an absorption model is evaluated to yield an absorption value for each projection pixel. Projection of a moving spline structure is fast and accurate. Projections of size 640 × 480 can be generated within 254 ms. Reconstructions using the projections show errors below 1 HU with a sharp reconstruction kernel. Traditional GPU-based acceleration schemes are not suitable for our reconstruction task. Even in the absence of noise, they result in errors up to 9 HU on average, although projection images appear to be correct under visual examination. Projections generated with our new method are suitable for the validation of novel CT reconstruction algorithms. For complex simulations, such as the evaluation of motion-compensated reconstruction algorithms, this kind of x-ray simulation will reduce the computation time dramatically.
The software and source code that was created for this paper can be found at ![]() http://conrad.stanford.edu/.
http://conrad.stanford.edu/.
Generating 3-D reconstructions of cardiac vasculature from angiographic C-arm CT (rotational angiography) data is a challenging problem. Currently, many approaches depend on a reconstruction from ECG-gated projection data either as a reference for further processing or as the final result. Due to imperfect gating, e.g. caused by irregular heart movement, residual motion corrupts these reconstructions. We present an algorithm to compensate for this residual motion. The approach is based on a deformable 2-D–2-D registration between the acquired projection data and a forward projection of the initial ECG-gated reconstruction. It does not depend on an explicit segmentation of vasculature or markers, and works without user interaction. The estimated 2-D deformation field is compensated for in the backprojection step of a subsequent reconstruction. The algorithm is evaluated on six clinical datasets, showing a clear decrease in artefact level and better visibility of structure in the compensated reconstructions.
 |
Many scientists in the field of x-ray imaging rely on the simulation of x-ray images. As the phantom models become more and more realistic, their projection requires high computational effort. Since x-ray images are based on transmission, many standard graphics acceleration algorithms cannot be applied to this task. However, if adapted properly, the simulation speed can be increased dramatically using state-of-the-art graphics hardware. A custom graphics pipeline that simulates transmission projections for tomographic reconstruction was implemented based on moving spline surface models. All steps from tessellation of the splines, projection onto the detector and drawing are implemented in OpenCL. We introduced a special append buffer for increased performance in order to store the intersections with the scene for every ray. Intersections are then sorted and resolved to materials. Lastly, an absorption model is evaluated to yield an absorption value for each projection pixel. Projection of a moving spline structure is fast and accurate. Projections of size 640 × 480 can be generated within 254 ms. Reconstructions using the projections show errors below 1 HU with a sharp reconstruction kernel. Traditional GPU-based acceleration schemes are not suitable for our reconstruction task. Even in the absence of noise, they result in errors up to 9 HU on average, although projection images appear to be correct under visual examination. Projections generated with our new method are suitable for the validation of novel CT reconstruction algorithms. For complex simulations, such as the evaluation of motion-compensated reconstruction algorithms, this kind of x-ray simulation will reduce the computation time dramatically.
The software and source code that was created for this paper can be found at ![]() http://conrad.stanford.edu/.
http://conrad.stanford.edu/.
 |
Coronary heart disease is the most common cause of death in the developed world. Its main cause is the partial or full ischemia of the coronary arteries. Today, the evaluation of theses stenoses in the interventional lab is performed using C-arm fluoroscopy.
A full 3-D visualisation can improve the clinical assessment. Currently, this may be done by an additional CT scan. But ideally, 3-D imaging should be performed in the interventional suite. This saves the additional diagnostic procedure and its associated costs and delivers information just in time.
Unfortunately, 3-D reconstruction of the coronary tree from C-arm data is a mathematically ill-posed problem and thus a difficult task to solve. Due to the long acquisition time of about five seconds, heart motion blurs the resulting image. There is previous work on the estimation of and compensation for the coronary vessel movement [1]. It delivers a good visualisation of the morphology of the coronary vessel tree. However, heart motion estimation is only approximate, leading to e.g. uncertainties in the quantitative determination of vessel diameters.
[1] C. Rohkohl, G. Lauritsch, L. Biller, M. Prümmer, J. Boese, J. Hornegger. Interventional 4D motion estimation and reconstruction of cardiac vasculature without motion periodicity assumption. Medical lmage Analysis, Vol. 14, pp. 687-694, 2010.
Many scientists in the field of x-ray imaging rely on the simulation of x-ray images. As the phantom models become more and more realistic, their projection requires high computational effort. Since x-ray images are based on transmission, many standard graphics acceleration algorithms cannot be applied to this task. However, if adapted properly, the simulation speed can be increased dramatically using state-of-the-art graphics hardware. A custom graphics pipeline that simulates transmission projections for tomographic reconstruction was implemented based on moving spline surface models. All steps from tessellation of the splines, projection onto the detector and drawing are implemented in OpenCL. We introduced a special append buffer for increased performance in order to store the intersections with the scene for every ray. Intersections are then sorted and resolved to materials. Lastly, an absorption model is evaluated to yield an absorption value for each projection pixel. Projection of a moving spline structure is fast and accurate. Projections of size 640 × 480 can be generated within 254 ms. Reconstructions using the projections show errors below 1 HU with a sharp reconstruction kernel. Traditional GPU-based acceleration schemes are not suitable for our reconstruction task. Even in the absence of noise, they result in errors up to 9 HU on average, although projection images appear to be correct under visual examination. Projections generated with our new method are suitable for the validation of novel CT reconstruction algorithms. For complex simulations, such as the evaluation of motion-compensated reconstruction algorithms, this kind of x-ray simulation will reduce the computation time dramatically.
The software and source code that was created for this paper can be found at ![]() http://conrad.stanford.edu/.
http://conrad.stanford.edu/.
 |
Generating 3-D reconstructions of cardiac vasculature from angiographic C-arm CT (rotational angiography) data is a challenging problem. Currently, many approaches depend on a reconstruction from ECG-gated projection data either as a reference for further processing or as the final result. Due to imperfect gating, e.g. caused by irregular heart movement, residual motion corrupts these reconstructions. We present an algorithm to compensate for this residual motion. The approach is based on a deformable 2-D–2-D registration between the acquired projection data and a forward projection of the initial ECG-gated reconstruction. It does not depend on an explicit segmentation of vasculature or markers, and works without user interaction. The estimated 2-D deformation field is compensated for in the backprojection step of a subsequent reconstruction. The algorithm is evaluated on six clinical datasets, showing a clear decrease in artefact level and better visibility of structure in the compensated reconstructions.
 |
High-density objects, like catheters, pacemakers or even contrast agent-filled vessels, cause characteristic streak artifacts in computed tomography (CT). Similar to metal artifacts, these streaks can be reduced by removing the dense object using segmentation and interpolation. First, we compare state-of-the-art interpolation methods like linear, spline and higher-order methods to the Healing Brush technique. Second, a new method is presented, that extracts a low-frequency model of the dense object and restores the decomposed X-ray intensity of the remaining tissue. This method is henceforth called Subtract-and-Shift. Compared to standard interpolation methods, it retains the measured structure that is superimposed and dominated by the dense object. The extracted structure is then used to replace the segmented pixel intensities of the object. The introduced method is compared to state-of-the-art interpolation methods using in-vivo data. First preliminary results show that Subtract-and-Shift can be superior to these interpolation methods.
 |