Room: 10.136

Sulaiman Vesal M. Sc.
Researcher in the Learning Approaches for Medical Big Data Analysis (LAMBDA) group at the Pattern Recognition Lab of the Friedrich-Alexander-Universität Erlangen-Nürnberg
Telephone: +49 1521 02 28519
E-mail: Sulaiman.vesal(at)fau.de
- Multi-modality breast image analysis and fusion
- Computer-aided detection and diagnosis of breast cancer
- Lesion classification in 3D MRI
- Quantitative analysis of breast imaging modalities
Large-scale breast image screening and analysis
Sulaiman Vesal, Nishant Ravikumar, AmirAbbas Davari, Stephan Ellmann, Andreas Maier
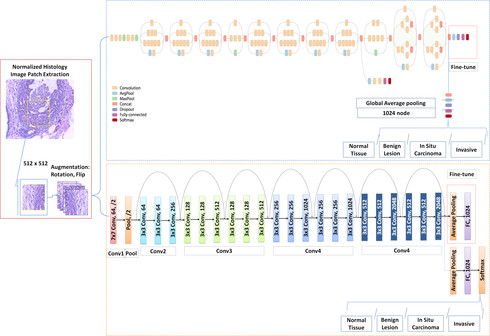
Breast cancer is one of the leading causes of mortality in women. Early detection and treatment are imperative for improving survival rates, which have steadily increased in recent years as a result of more sophisticated computer-aided-diagnosis (CAD) systems. CAD systems are essential to reduce subjectivity and supplement the analyses conducted by specialists. We propose a transfer learning-based approach, for the task of breast histology image classification into four tissue sub-types, namely, normal, benign, Insitu carcinoma and invasive carcinoma. The histology images, provided as part of the BACH 2018 grand challenge, were first normalized to correct for color variations induced during slide preparation. Subsequently, image patches were extracted and used to fine-tune Google`s Inception-V3 and ResNet50 convolutional neural networks, both pre-trained on the ImageNet database, enabling them to learn domain-specific features, necessary to classify the histology images. Classification accuracy was evaluated using 3-folds. The Inception-V3 network achieved an average test accuracy of 97.08\% for four classes, marginally outperforming the ResNet50 network, which achieved an average accuracy of 96.66\%.
Sulaiman Vesal, Nishant Ravikumar, Andreas Maier; MIC/NSS 2018
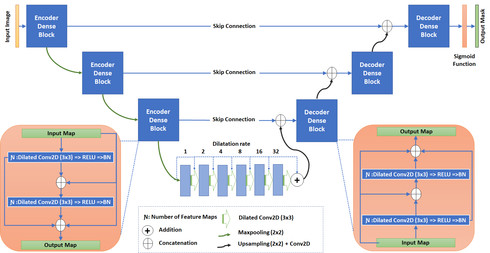
There has been a steady increase in the incidence of skin cancer worldwide, with a high rate of mortality. Early detection and segmentation of skin lesions is crucial for timely diagnosis and treatment, necessary to improve the survival rate of patients. However, skin lesion segmentation is a challenging task due to the low contrast of lesions and their high similarity in terms of appearance, to healthy tissue. This underlines the need for an accurate and automatic approach for skin lesion segmentation. To tackle this issue, we propose a convolutional neural network (CNN) called SkinNet.
The proposed CNN is a modified version of U-Net. We compared the performance of our approach with other state-of-the-art techniques, using the ISBI 2017 challenge dataset. Our approach outperformed the others in terms of the Dice coefficient, Jaccard index and sensitivity, evaluated on the held-out challenge test data set, across 5-fold cross validation experiments. SkinNet achieved an average value of 85.10, 76.67 and 93\%, for the DC, JI and SE, respectively.
Sulaiman Vesal, Nishant Ravikumar, Andreas Maier; SATCOM-MICCAI 2018
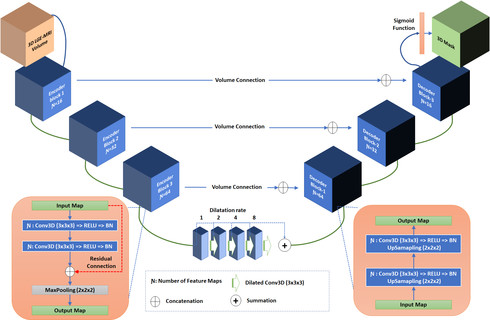
Segmentation of the left atrial chamber and assessing its morphology, are essential for improving our understanding of atrial fibrillation, the most common type of cardiac arrhythmia. Automation of this process in 3D gadolinium enhanced-MRI (GE-MRI) data is desirable, as manual delineation is time-consuming, challenging and observer-dependent. Recently, deep convolutional neural networks (CNNs) have gained tremendous traction and achieved state-of-the-art results in medical image segmentation. However, it is difficult to incorporate local and global information without using contracting (pooling) layers, which in turn reduces segmentation accuracy for smaller structures. In this paper, we propose a 3D CNN for volumetric segmentation of the left atrial chamber in LGE-MRI. Our network is based on the well known U-Net architecture. We employ a 3D fully convolutional network, with dilated convolutions in the lowest level of the network, and residual connections between encoder blocks to incorporate local and global knowledge. The results show that including global context through the use of dilated convolutions, helps in domain adaptation, and the overall segmentation accuracy is improved in comparison to a 3D U-Net.
Sulaiman Vesal, Shreyas Malakarjun Patil, Nishant Ravikumar,and Andreas K. Maier; ISIC-MICCAI 2018
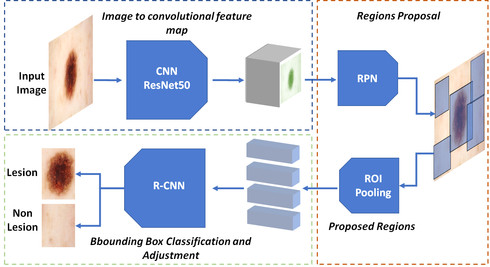
Early detection and segmentation of skin lesions is crucial for timely diagnosis and treatment, necessary to improve the survival rate of patients. However, manual delineation is time consuming and subject to intra- and inter-observer variations among dermatologists. This underlines the need for an accurate and automatic approach to skin lesion segmentation. To tackle this issue, we propose a multi-task convolutional neural network (CNN) based, joint detection and segmentation frame-work, designed to initially localize the lesion and subsequently, segment it. A ‘Faster region-based convolutional neural network’ (Faster-RCNN)which comprises a region proposal network (RPN), is used to generate bounding boxes/region proposals, for lesion localization in each image.The proposed regions are subsequently refined using a softmax classifier and a bounding-box regressor. The refined bounding boxes are finally cropped and segmented using ‘SkinNet’, a modified version of U-Net.We trained and evaluated the performance of our network, using the ISBI 2017 challenge and the PH2 datasets, and compared it with the state-of-the-art, using the official test data released as part of the challenge for the former. Our approach outperformed others in terms of Dice coefficients (>0.93), Jaccard index (>0.88), accuracy (>0.96) and sensitivity (>0.95), across five-fold cross validation experiments.
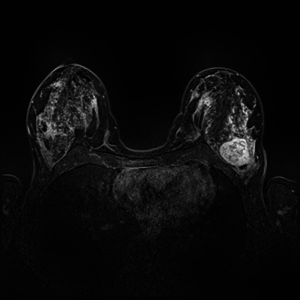
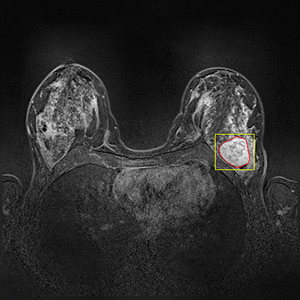
Dynamic Contrast Enhanced Magnetic Resonance Imaging (DCE-MRI) recently proved to be quite helpful in screening high-risk women and in staging newly diagnosed breast cancer patients. Accurate segmentation of tumor lesion and selection of suspicious regions of interest (ROIs) is a critical pre-processing step in DCE-MRI data evaluation. The goal of this project is to develop and evaluate methods for automatic and semi-automatic detection of suspicious ROIs for breast DCE-MRI, segmentation of tumor lesions, morphological feature extraction and classification of tumors in terms of benign and malignant. I will try to use deep learning algorithms as state-of-the-art approaches for every possible stages. This project is partly connected to Big-Thera project and the patients data are provided by Radiology department of Killinikum University of Erlangen.

 +49 9131 85 27799
+49 9131 85 27799
 +49 9131 85 27270
+49 9131 85 27270