
Friedrich-Alexander-Universität Erlangen
Lehrstuhl für Mustererkennung
Martensstraße 3
91058 Erlangen

Rigid, non-rigid and multi-modal 3-D surface registration.

The recent introduction of low-cost devices for real-time acquisition of dense 3-D range imaging (RI) streams has attracted a great deal of attention. Many research communities will benefit from this evolution. However, to date, there exists no open source framework that is explicitly dedicated to real-time processing of RI streams.
In this paper, we present the Range Imaging Toolkit (RITK). The goal is to provide a powerful yet intuitive software platform that facilitates the development of range image stream applications. In addition to its usage as a library within existing software, the toolkit supports developers with an easy-to-use development and rapid prototyping infrastructure for the creation of application-specific RITK modules or standalone solutions. Putting a strong focus on modularity, this application-layer allows for distribution and reuse of existing modules. In particular, a pool of dynamically loadable plugins can be assembled into tailored RI processing pipelines during runtime. RITK puts emphasis on real-time processing of range image streams and proposes the use of a dedicated pipeline mechanism. Furthermore, we introduce a powerful and convenient interface for range image processing on the graphics processing unit (GPU). Being designed thoroughly and in a generic manner, the toolkit is able to cope with the broad diversity of data streams provided by available RI devices and can easily be extended by custom range imaging sensors or processing modules.
RITK is an open source project and is publicly available under![]() www.cs.fau.de/ritk.
www.cs.fau.de/ritk.

Recent advances in range imaging (RI) have enabled dense 3-D scene acquisition in real-time. However, due to physical limitations and the underlying range sampling principles, range data are subject to noise and may contain invalid measurements. Hence, data preprocessing is a prerequisite for practical applications but poses a challenge with respect to real-time constraints.
In this paper, we propose a generic and modality-independent pipeline for efficient RI data preprocessing on the graphics processing unit (GPU). The contributions of this work are efficient GPU implementations of normalized convolution for the restoration of invalid measurements, bilateral temporal averaging for dynamic scenes, and guided filtering for edge-preserving denoising. Furthermore, we show that the transformation from range measurements to 3-D world coordinates can be computed efficiently on the GPU. The pipeline has been evaluated on real data from a Time-of-Flight sensor and Microsoft's Kinect. In a run-time performance study, we show that for VGA-resolution data, our preprocessing pipeline runs at ~100 fps on an off-the-shelf consumer GPU.
 |
 |
 |
 |
Time-of-Flight (ToF) imaging is a promising technology for real-time metric surface acquisition and has recently been proposed for a variety of medical applications such as intra-operative organ surface registration. However, due to limitations of the sensor, range data from ToF cameras are subject to noise and contain invalid outliers. While systematic errors can be eliminated by calibration, denoising provides the fundamental basis to produce steady and reliable surface data.
In this paper, we introduce a generic framework for high-performance ToF preprocessing that satisfies real-time constraints in a medical environment. The contribution of this work is threefold. First, we address the restoration of invalid measurements that typically occur with specular reflections on wet organ surfaces. Second, we compare the conventional bilateral filter with the recently introduced concept of guided image filtering for edge preserving denoising. Third, we have implemented the pipeline on the graphics processing unit (GPU), enabling high-quality preprocessing in real-time. In experiments, the framework achieved a depth accuracy of 0.8 mm (1.4 mm) on synthetic (real) data, at a total runtime of 40 ms.
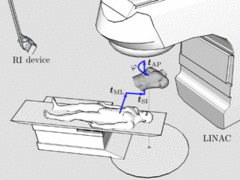
In radiation therapy, prior to each treatment fraction, the patient must be aligned to computed tomography (CT) data. Patient setup verification systems based on range imaging (RI) can accurately verify the patient position and adjust the treatment table at a fine scale, but require an initial manual setup using lasers and skin markers. We propose a novel markerless solution that enables a fully-automatic initial coarse patient setup. The table transformation that brings template and reference data in congruence is estimated from point correspondences based on matching local surface descriptors. Inherently, this point-based registration approach is capable of coping with gross initial misalignments and partial matching. Facing the challenge of multi-modal surface registration (RI/CT), we have adapted state-of-the-art descriptors to achieve invariance to mesh resolution and robustness to variations in topology. In a case study on real data from a low-cost RI device (Microsoft Kinect), the performance of different descriptors is evaluated on anthropomorphic phantoms. Furthermore, we have investigated the system's resilience to deformations for mono-modal RI/RI registration of data from healthy volunteers. Under gross initial misalignments, our method resulted in an average angular error of 1.5° and an average translational error of 13.4 mm in RI/CT registration. This coarse patient setup provides a feasible initialization for subsequent refinement with verification systems.
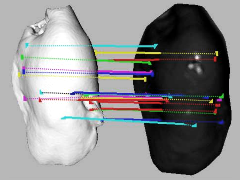
In the field of image-guided liver surgery (IGLS), the initial registration of the intraoperative organ surface with preoperative tomographic image data is performed on manually selected anatomical landmarks. In this paper, we introduce a fully automatic scheme that is able to estimate the transformation for initial organ registration in a multi-modal setup aligning intraoperative time-of-flight (ToF) with preoperative computed tomography (CT) data, without manual interaction. The method consists of three stages: First, we extract geometric features that encode the local surface topology in a discriminative manner based on a novel gradient operator. Second, based on these features, point correspondences are established and deployed for estimating a coarse initial transformation. Third, we apply a conventional iterative closest point (ICP) algorithm to refine the alignment. The proposed method was evaluated for an open abdominal hepatic surgery scenario with in-vitro experiments on four porcine livers. The method achieved a mean distance of 4.82 ± 0.79 mm and 1.70 ± 0.36 mm for the coarse and fine registration, respectively.

Methods for 2D/3D face recognition typically combine results obtained independently from the 2D and 3D data, respectively. There has not been much emphasis on data fusion at an early stage, even though it is at least potentially more powerful to exploit possible synergies between the two modalities. In this paper, we propose photogeometric features that interpret both the photometric texture and geometric shape information of 2D manifolds in a consistent manner. The 4D features encode the spatial distribution of gradients that are derived generically for any scalar field on arbitrary organized surface meshes. We apply the descriptor for biometric face recognition with a time-of-flight sensor. The method consists of three stages: (i) facial landmark localization with a HOG/SVM sliding window framework, (ii) extraction of photogeometric feature descriptors from time-of-flight data, using the inherent grayscale intensity information of the sensor as the 2D manifold's scalar field, (iii) probe matching against the gallery. Recognition based on the photogeometric features achieved 97.5% rank-1 identification rate on a comprehensive time-of-flight dataset (26 subjects, 364 facial images).

The modeling of three-dimensional scene geometry from temporal point cloud streams is of particular interest for a variety of computer vision applications. With the advent of RGB-D imaging devices that deliver dense, metric and textured 6-D data in real-time, on-the-fly reconstruction of static environments has come into reach. In this paper, we propose a system for real-time point cloud mapping based on an efficient implementation of the iterative closest point (ICP) algorithm on the graphics processing unit (GPU). In order to achieve robust mappings at real-time performance, our nearest neighbor search evaluates both geometric and photometric information in a direct manner. For acceleration of the search space traversal, we exploit the inherent computing parallelism of GPUs. In this work, we have investigated the fitness of the random ball cover (RBC) data structure and search algorithm, originally proposed for high-dimensional problems, for 6-D data. In particular, we introduce a scheme that enables both fast RBC construction and queries. The proposed system is validated on an indoor scene modeling scenario. For dense data from the Microsoft Kinect sensor (640x480 px), our implementation achieved ICP runtimes of < 20 ms on an off-the-shelf consumer GPU.

In radiation therapy, studies have shown that external body motion correlates with the internal tumor position. Based on a patient-specific model learnt prior to the first fraction, the tumor location can be inferred from an external surrogate during treatment, superseding intra-fractional radiographic imaging. Existing solutions rely on a single 1-D respiratory surrogate and exhibit a limited level of accuracy due to inter-cycle variation. Recent work indicated that considering multiple regions would yield an enhanced model. Hence, we propose a novel solution based on a single-shot active triangulation sensor that acquires a sparse grid of 3D measurement lines in real time, using two perpendicular laser line pattern projection systems. Building on non-rigid point cloud registration, the elastic displacement field representing the torso deformation with respect to a reference is recovered. This displacement field can then be used for model-based tumor tracking. We present measurement results and non-rigid displacement fields.
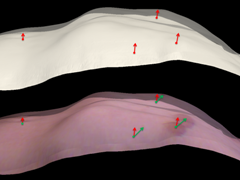
In radiation therapy, the estimation of torso deformations due to respiratory motion is an essential component for real-time tumor tracking solutions. Using range imaging (RI) sensors for continuous monitoring during the treatment, the 3-D surface motion field is reconstructed by a non-rigid registration of the patient’s instantaneous body surface to a reference. Typically, surface registration approaches rely on the pure topology of the target. However, for RI modalities that additionally capture photometric data, we expect the registration to benefit from incorporating this secondary source of information. Hence, in this paper, we propose a method for the estimation of 3-D surface motion fields using an optical flow framework in the 2-D photometric domain. In experiments on real data from healthy volunteers, our photometric method outperformed a geometry-driven surface registration by 6.5% and 22.5% for normal and deep thoracic breathing, respectively. Both the qualitative and quantitative results indicate that the incorporation of photometric information provides a more realistic deformation estimation regarding the human respiratory system.

The management of intra-fractional respiratory motion is becoming increasingly important in radiation therapy. Based on in advance acquired accurate 3D CT data and intra-fractionally recorded noisy Time-of-Flight (ToF) range data an improved treatment can be achieved. In this paper, a variational approach for the joint registration of the thorax surface extracted from a CT and a ToF image and the denoising of the ToF image is proposed. This enables a robust intra-fractional full torso surface acquisition and deformation tracking to cope with variations in patient pose and respiratory motion. Thereby, the aim is to improve radiotherapy for patients with thoracic, abdominal and pelvic tumors. The approach combines a Huber norm type regularization of the ToF data and a geometrically consistent treatment of the shape mismatch. The algorithm is tested and validated on synthetic and real ToF/CT data and then evaluated on real ToF data and 4D CT phantom experiments.